Precision detection and quantification of nucleic acid (DNA / RNA) molecules using digital PCR analysis
Digital PCR analysis is a highly sensitive method for detecting and precisely quantifying nucleic acid (DNA / RNA) molecules. Unlike qPCR, dPCR can apply multiple partitions, enabling absolute quantification of your sample. In addition to exquisite accuracy, this method is also highly reproducible and significantly reduces potential interference from substances within the sample matrix (the matrix effect) by isolating and minimizing the impact of inhibitor reactions.
Researchers typically leverage digital PCR services to drive advancements in cell and gene therapy, biomarker discovery, and RNA/DNA analysis.
Common applications of dPCR analysis
- Cell and gene therapy: Biodistribution in complex matrix
- Recombinant vector integrity
- Gene expression analysis: Monitoring subtle changes that cannot be detected by qPCR
- Detection of rare sequences in cancer (detection of mutation below levels detectable by current tests in wild background context)
- Liquid Biopsy: Circulating nucleic acids (cfDNA) & circulating tumor cells (ctDNA) measurement
- Hypermethylation DNA / splicing analysis / KO efficiency
- Copy number variation (CNV): 1.2-fold change, Allelic variants (SNPs), KO efficiency / genome edit detection
- Pathogen detection / microbiome / viral load analysis

Partitioning in digital PCR analysis
One of the key strengths of digital PCR lies in its partitioning capability, which enhances sensitivity and precision in quantifying nucleic acids.
Process overview:
- Partitioning the reaction: The PCR reaction mixture is divided into thousands of individual micro-reactions (partitions).
- Template distribution: DNA or RNA templates are randomly distributed across these partitions, with some containing target molecules and others not.
- End-point amplification: PCR amplification occurs independently in each partition to the end-point.
- Fluorescence detection: Fluorescent probes such as TaqMan or SYBR Green are used to detect positive partitions. Multiplexing allows for the simultaneous detection of multiple targets.
- Quantification via Poisson statistics: By analyzing the proportion of positive partitions, Poisson statistics are applied to accurately determine the absolute quantity of target nucleic acids per well.
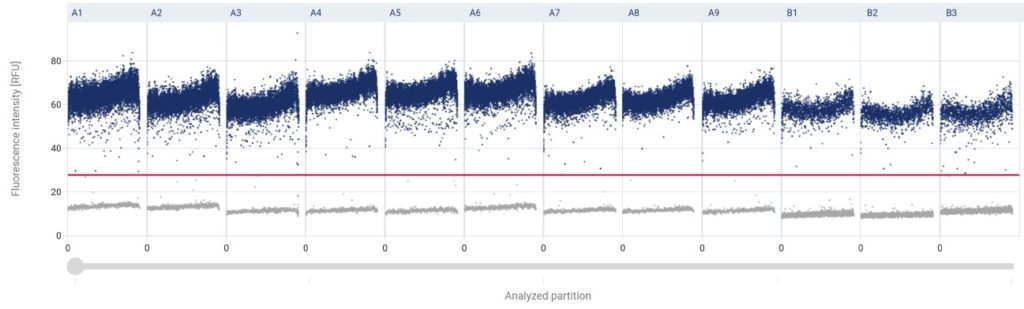
(Above) Illustration of results following digital PCR analysis. In blue: positive partitions (containing DNA or RNA). In grey: negative partitions. Each column corresponds to one dPCR well. The vertical axis corresponds to fluorescence level in each partition measured at the end of the dPCR run.
Digital PCR vs qPCR: Which service best suits your requirement?
| qPCR | dPCR | |
| Quantification | Relative: Requires a well characterized standard curve | Absolute: Standard curve not required |
| Precision | Low | High |
| Sensitivity | High | Very high |
| Measurement | Real-time: Impacted by PCR efficiency | End point: Not impacted by PCR efficiency |
| Enzymatic reaction | In the entire PCR well All sample / well Precision and Sensitivity: High but lower than dPCR |
Partitioning in 8,500 or 26,000 micro-reactions 1 molecule / partition Precision and sensitivity: High |
| Sensitivity to inhibitors | High | Low |
| Multiplex capabilities | Up to 6 plex | Up to 12 plex |
| Dynamic range | ~7 log | ~5 log
Sample dilution for high target concentrations |
| Reactions / plate | 96 or 384 wells | 24 (8,500 or 26,000 partitions) or 96 wells
(8,500 partitions) |
Advanced bioanalysis services
Oncodesign Services offers extensive expertise in bioanalysis and a large portfolio of bioanalytical technologies, including:
- Qiagen QIAcuity digital PCR (dPCR)
- ThermoFisher QuantStudio 7 Flex Real-Time PCR System (qPCR)
- LC-UV, LC-MS and LC-HRMS
- Flow cytometry (FACS)
- Ligand Binding assays
- Immunogenicity assays
Our experienced team can facilitate method development, method transfer, method validation, and sample analysis (discovery, GLP and GcLP-grade studies), as well as providing guidance around the best approaches and methodologies to meet your specific research requirements.