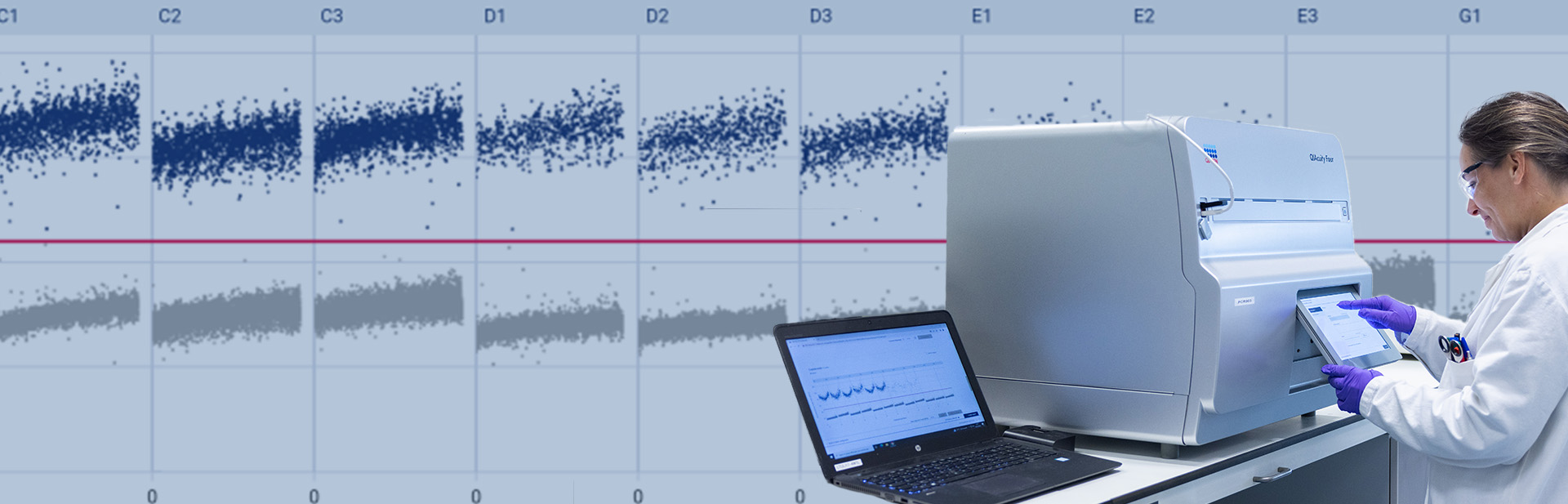


PCR is a laboratory technique that was made very popular during the COVID-19 pandemic. The main principle, developed in the 1980s, comes from the amplification of a sample to make it quantifiable.
The first approach coining the term digital PCR (dPCR) was developed in 19991, and various instruments began to arrive on the scene around 2010. At Oncodesign Services, we have implemented the chamber-based technique (QIAcuity Four Digital PCR System from Qiagen) in order to better respond to our customers’ demands in several environments: discovery (R&D) and regulatory (GLP and GcLP).
What is PCR?
It stands for Polymerase Chain Reaction. This enzymatic reaction allows for the specific amplification of a nucleic acid sequence (DNA or RNA) that is not abundant and therefore neither detectable nor quantifiable without this amplification.
It is a very powerful tool in molecular biology and has many applications, including genetic research, medical diagnostics, and forensic analysis.
Here we look at why you should switch to dPCR over traditional quantitative PCR (qPCR).
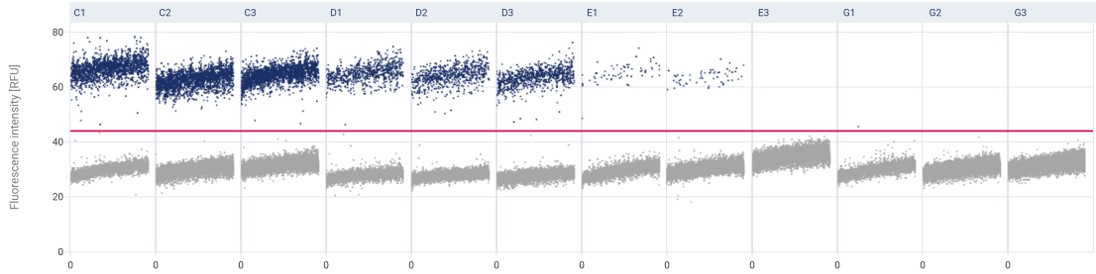
1. Increased precision and accuracy
dPCR partitions a sample into many smaller reactions, which reduces the variability between samples and increases the precision and accuracy of the measurement. This makes it easier to detect rare mutations or subtle changes in gene expression levels.
2. Decreased matrix effect
Thanks to sample partitioning, PCR inhibitors are diluted into each partition, which drastically mitigates matrix effect. This makes dPCR particularly useful for complex matrices like feces or tissues.
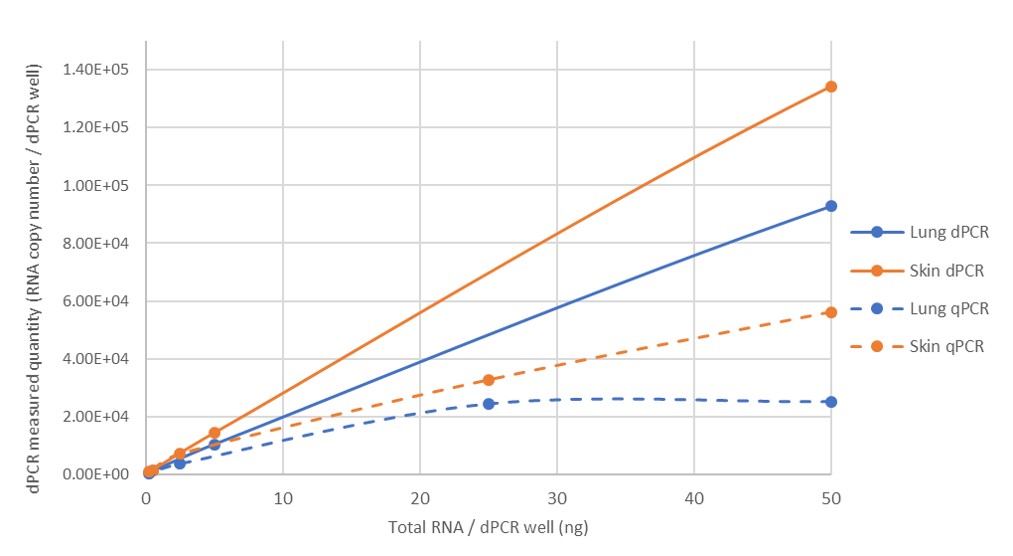
3. Higher sensitivity
dPCR is able to detect smaller amounts of target molecules in a sample compared to traditional qPCR, making it useful for applications where very low amounts of DNA or RNA need to be detected.
4. Reduced reliance on standard curves
Traditional qPCR requires a standard curve to calibrate measurements, which can introduce errors and variability. dPCR counts the number of positive reactions, allowing for absolute quantification without the need of a standard curve.
5. Robustness
By utilizing a high number of small partitions of very small volumes, dPCR is less susceptible to the effects of inhibitors and variations in sample preparation, making it more robust and reliable for quantitative measurements.
Various application of dPCR…
The main applications typically involve developing and validating methods to support biodistribution/shedding studies in the context of IND-enabling programs. Hence, Oncodesign Services propose development, qualification and/or GLP compliant validation of dPCR methods for quantification of gene therapy products in many tissues / fluids from different species.
Another application in oncology concerns the analysis of DNA methylation. DNA methylation is an epigenetic modification that consists of a modification on the cytosine of CpG dinucleotides (CpG islands). Generally speaking, when the promoter of a gene is methylated, the expression of the downstream gene is repressed. This modification can therefore influence gene expression and is a point of interest in oncology. The identification of specific hypermethylated genes is indeed a biomarker in different types of cancers.
Oncodesign Services develop also bacteria and virus detection by dPCR (from the design of primers/probe to sample assays) in different type of matrices.
Last but not least, dPCR, recognized as one of the most suitable approaches for rare event detection, will facilitate the use of circulating-tumor DNA fragments (ctDNA) as potential predictor biomarkers in clinical practice
About the author:
The article was written with Dr. Sophie Champlot. In her current role at Oncodesign Services, Sophie is GLP-GcLP Study Director in the DMPK-Bioanalytical Sciences Department, and specialized in molecular biology/qPCR/dPCR to satisfy customers’ needs.
Reference:
1 Vogelstein and Kinzler, 1999 – DOI: 10.1073/pnas.96.16.9236
Associated keywords:
dPCR / qPCR / bioanalysis /
